Small Ubiquitin-like Modifier (SUMO) refers to a family of small proteins that function as post-translational modifiers, primarily affecting the behavior and fate of target proteins. SUMOylation is evolutionarily conserved and found in all eukaryotic cells.
Classification and Structural Features
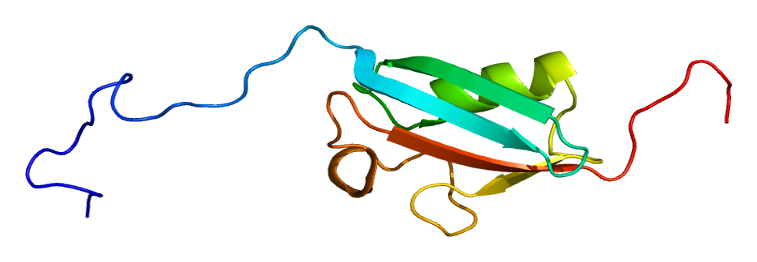
SUMO is a ubiquitin-like protein, sharing ~18% sequence similarity with ubiquitin and possessing the characteristic β-grasp fold (“ββαββαβ”).
A C-terminal diglycine motif (GG) is critical for conjugation to lysine residues of target proteins.
Unlike ubiquitin, SUMO proteins have an intrinsically disordered N-terminus (IDR)—13–23 amino acids—which modulates conformational flexibility and facilitates phase separation and dynamic protein interactions.
Humans have five SUMO paralogs:
SUMO1–3 are the most studied; SUMO2/3 share ~97% identity and often function interchangeably.
SUMO1 shares ~45% identity with SUMO2/3 and often terminates SUMO chains.
SUMO4–5 are less characterized.
Yeast (Saccharomyces cerevisiae) expresses only a single SUMO gene: SMT3.
Mechanism of SUMOylation
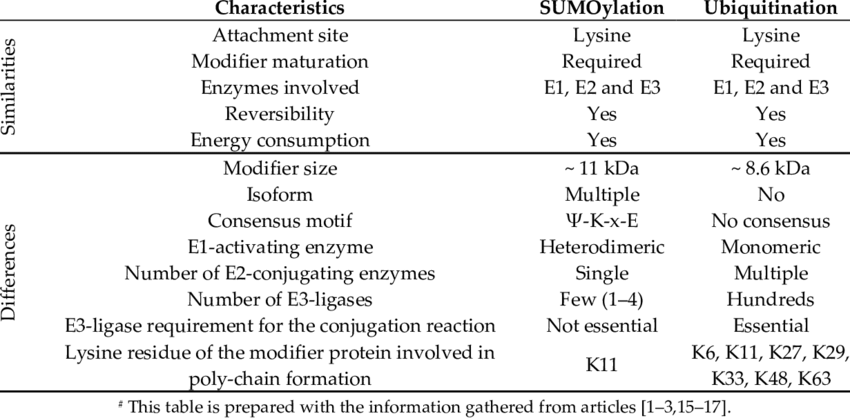
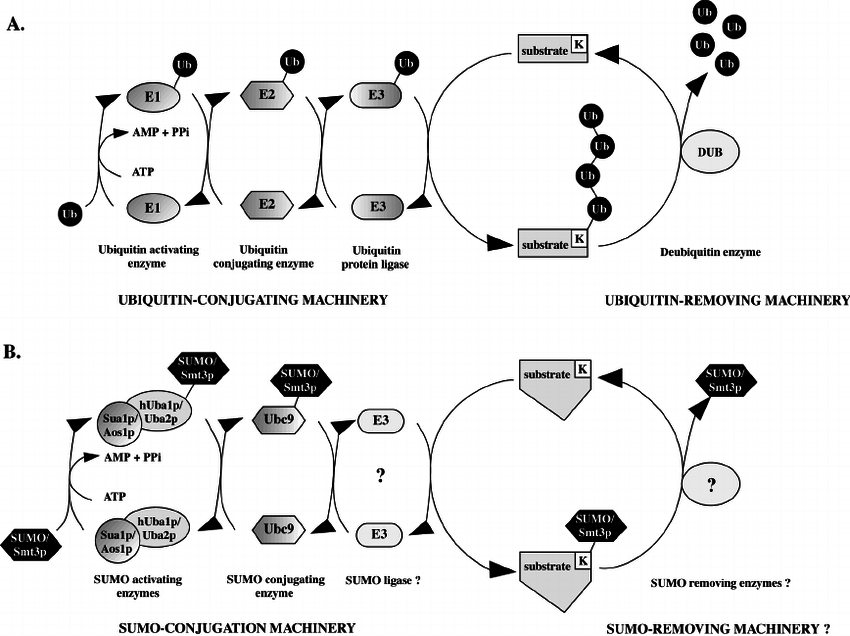
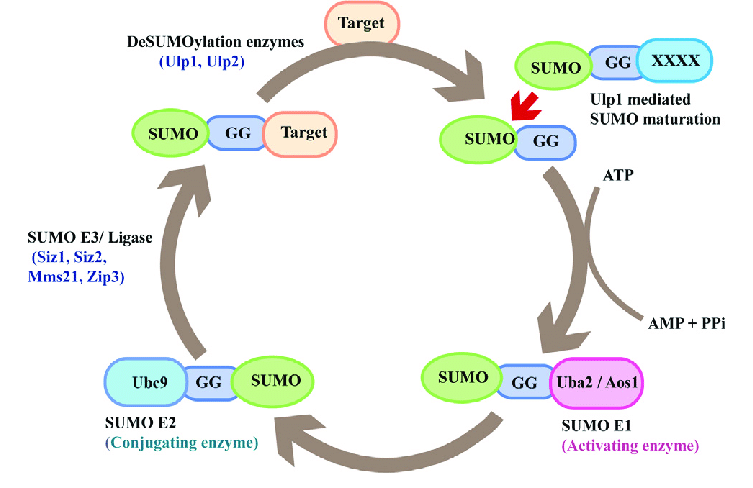
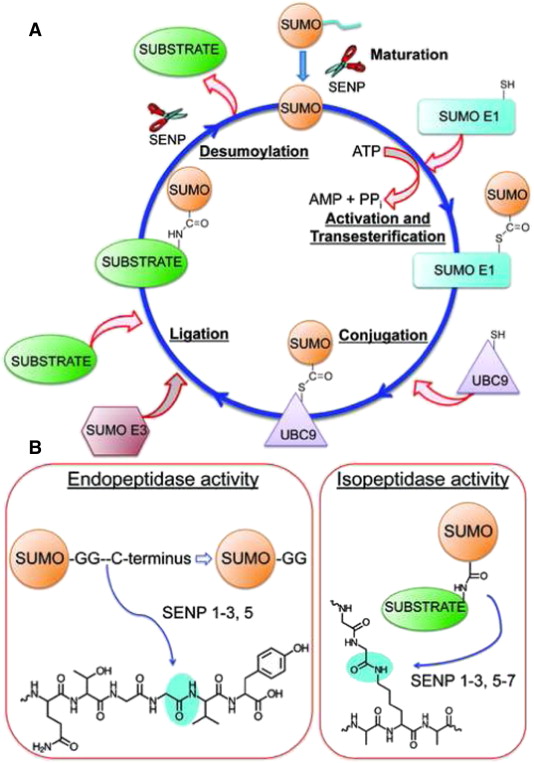
Sumoylation is a reversible, covalent post-translational modification that occurs through a three-enzyme cascade:
Maturation: SUMO precursors are cleaved by SUMO-specific proteases to expose the terminal diglycine motif.
Activation (E1 enzyme): The mature SUMO is activated in an ATP-dependent manner by an E1 heterodimer (SAE1/SAE2 in humans).
Conjugation (E2 enzyme): Activated SUMO is transferred to the sole E2 conjugating enzyme Ubc9, which recognizes the SUMO consensus motif ψKx(D/E) on target proteins.
Ligation (E3 ligases): E3 ligases increase substrate specificity and efficiency, forming an isopeptide bond between SUMO and the lysine residue of the target protein.
Types of SUMOylation: Mono-, Multi-, and PolySUMOylation
- MonoSUMOylation: Involves attachment of a single SUMO unit; typically regulates protein localization and interaction.
PolySUMOylation: SUMO2/3 form polymeric chains through K11 of SUMO itself.
SUMO1 usually terminates SUMO chains.
These modifications modulate:
Protein stability
Nuclear-cytoplasmic transport
Condensate assembly
Recruitment of ubiquitin ligases (STUbLs)
DeSUMOylation
SUMOylation is reversed by SUMO-specific proteases:
In yeast: Ulp1 (essential) and Ulp2/Smt4 (nonessential, nuclear).
In humans: SENP1–SENP7 regulate SUMO turnover.
SENP6/7 preferentially act on SUMO2/3 polySUMO chains.
Functions and Biological Roles
Sumoylation regulates a wide array of cellular processes:
Transcriptional regulation
DNA replication and repair
Cell cycle control
Nuclear transport
Apoptosis
Stress responses (e.g., heat shock, oxidative stress, DNA damage)
Additionally:
SUMO-Interacting Motifs (SIMs) enable non-covalent SUMO–protein interactions, essential for:
Protein complex formation
Biomolecular condensates and LLPS (e.g., PML nuclear bodies)
Group SUMOylation: Entire protein complexes may be sumoylated together via recruited E3 ligases, enhancing coordinated regulation.
SUMO and Disease
Defective SUMO conjugation/deconjugation is implicated in:
Cancer: Altered sumoylation of transcription factors, tumor suppressors, and kinases.
Neurodegeneration: SUMOylation affects aggregation-prone proteins in Alzheimer’s and Huntington’s disease.
Cellular adaptation: In yeast, loss of Ulp2 leads to aneuploidy as a short-term adaptation, followed by compensatory genomic changes.
Regulation of SUMOylation by Other PTMs
SUMO itself can be modified:
Phosphorylation, acetylation, ubiquitination of SUMO or SUMOylated substrates affects specificity, localization, and stability.
Example: LKB1, a kinase regulating AMPK, is SUMOylated at K178 (SUMO1 or SUMO2 depending on cell context), modulating its nuclear-cytoplasmic shuttling.
K48 acetylation influences SUMOylation dynamics and LKB1’s interaction with STRADα, affecting tumor growth in liver cancer.
Alternative Splicing of SUMO Genes
SUMO1α, SUMO2α, and SUMO3α are alternative isoforms:
Some are non-conjugatable due to disrupted β-grasp fold.
They may still interact with SIM-containing proteins, affecting SUMO-dependent signaling indirectly.
SUMO3α retains conjugation ability but targets a distinct set of proteins.
SUMOylation in Drug Development: A Promising Therapeutic Avenue
SUMOylation, a reversible post-translational modification, has emerged as a compelling target in drug development due to its essential roles in diverse cellular processes and its dysregulation in numerous diseases, especially cancer and neurodegenerative disorders.
1. SUMOylation as a Therapeutic Target
Aberrations in the SUMOylation/deSUMOylation balance are implicated in a wide range of human diseases.
Many of these disorders, including cancer and Alzheimer’s disease, are associated with altered phase-separated condensates, suggesting a link between SUMOylation and biomolecular compartmentalization.
2. SUMOylation in Cancer Treatment
Oncogenic Mechanisms and Targets
Upregulation of SUMOylation is frequently observed in various cancers.
For instance, monoSUMOylation of MYC enhances its stability and transcriptional activity, promoting tumorigenesis and metastasis.
In hepatocellular carcinoma (HCC):
LKB1, a kinase with context-dependent roles in cancer, is SUMOylated at lysine 178 by SUMO-2, impeding its nuclear export.
This modification is linked to tumor growth and invasiveness, particularly under hypoxic conditions.
Upregulation of the E2 enzyme Ubc9 and E3 ligase CBX4 in HCC correlates with poor prognosis.
Therapeutic Approaches and SUMO Inhibitors
Global SUMOylation Inhibitors:
TAK-981: A clinical-stage SUMOylation inhibitor with anti-cancer and antiviral effects, partly by enhancing type I interferon (IFN) signaling.
ML-792: A potent E1 inhibitor that reduces cancer cell viability and MYC expression.
Natural Compounds: Ginkgolic acid and anacardic acid inhibit SUMOylation in vitro by targeting the E1 enzyme.
Pathway-Specific Targeting:
Arsenic trioxide in APL therapy promotes degradation of PML-RARA via a polySUMOylation- and STUbL-dependent pathway.
SENP Inhibitors: Peptide- and small molecule-based inhibitors of SENPs (e.g., SENP6, SENP7) may be promising in SUMO2-mediated oncogenic pathways like in HCC.
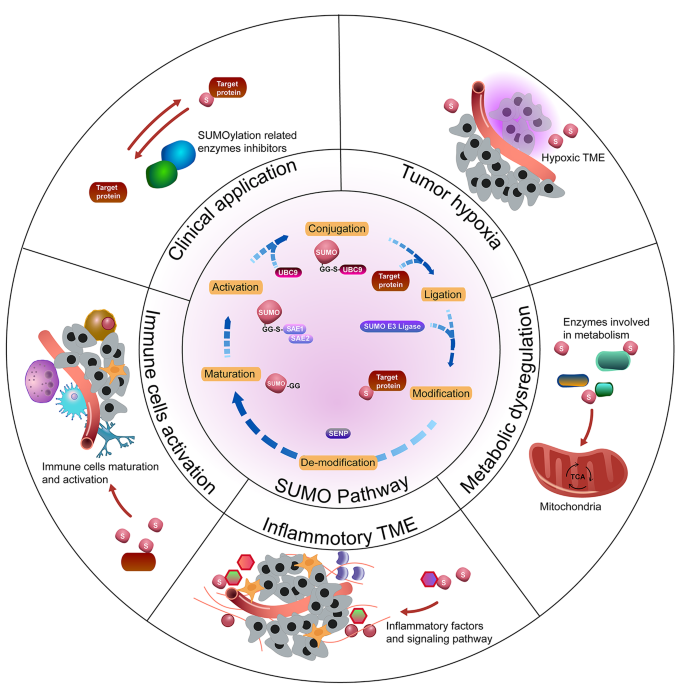
3. SUMOylation in Neurodegenerative Diseases
SUMOylation influences protein aggregation, a hallmark of diseases like Alzheimer’s, Parkinson’s, and Huntington’s.
Disease Mechanisms
In Alzheimer’s and Parkinson’s, monoSUMOylation of tau enhances its aggregation.
In Huntington’s disease:
SUMO1 modification stabilizes and solubilizes mutant huntingtin.
SUMO2 modification leads to the formation of insoluble aggregates, which are associated with disease progression.
Reducing global SUMOylation has shown potential in slowing neurodegeneration in experimental models.

4. Challenges and Considerations in SUMO-Targeted Therapy
Selectivity: Targeting specific oncogenic SUMOylation events is more desirable than broadly suppressing SUMOylation.
Paralogue Specificity:
Understanding the distinct functions of SUMO1 vs. SUMO2/3 is essential.
SUMO2, being more abundant in tumor cells, presents a more focused therapeutic target.
Alternative Isoforms:
SUMO alphas (SUMO1α, SUMO2α, SUMO3α) have unique properties and may regulate SUMO-related pathways through non-conjugatable interactions.
Cellular Adaptation:
Cells can adapt to SUMO dysregulation through genomic alterations, making long-term therapeutic strategies complex and necessitating deeper understanding of SUMO system plasticity.
In summary, SUMOylation plays a pivotal role in maintaining cellular homeostasis and regulating protein function. As our understanding deepens, it is becoming clear that SUMOylation is not just a regulatory add-on but a dynamic and essential process with broad implications for health and disease. Future research will likely reveal novel therapeutic opportunities by targeting this pathway.
Explore the latest research on SUMOylation here.