A groundbreaking study published in Nature Materials presents a computational framework for creating bifaceted protein nanomaterials—also known as Janus protein nanoparticles—with independently addressable faces. This approach overcomes the limitations of traditional symmetric protein designs, which preclude functional differentiation across particle surfaces.
1. Introduction & Design Rationale
Traditional nanoparticle designs rely on high symmetry and identical subunits, restricting anisotropic functionalization (i.e., functional diversity across different faces of the particle).
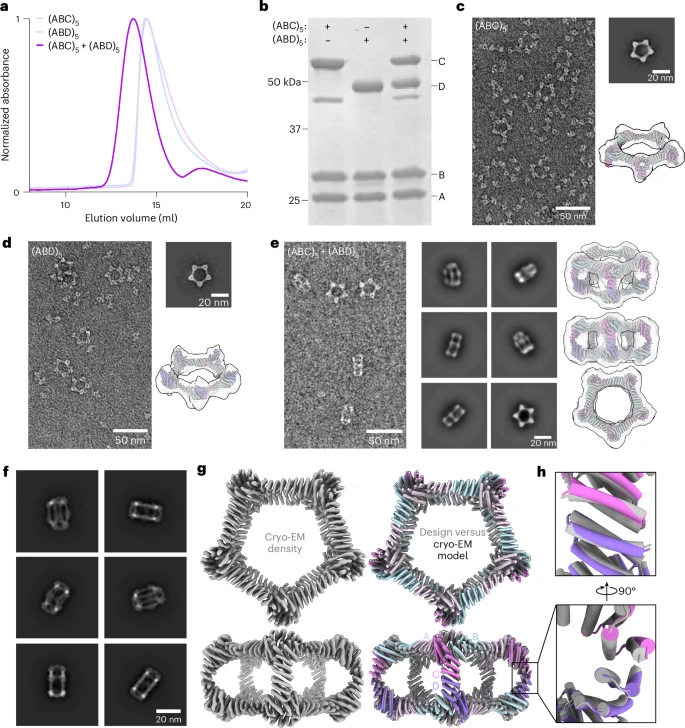
The authors introduce a novel pseudo‑D₅ (pD5) architecture—formulated as (ABC)₅–(ABD)₅—by designing an asymmetric interface between two heterotrimeric building blocks, enabling each of the six subunits to be uniquely functionalizable and independently addressable.
2. Computational Design Pipeline
Assembly Architecture
Starting from a 15-subunit “Crown_C5‑1” ring built from five heterotrimers (ABC), two such rings are docked along the five-fold axis to generate a 30‑subunit assembly with D₅ symmetry.
Asymmetric Interface Design
ProteinMPNN, a deep learning sequence-design tool, was used to specify a C–D asymmetric interface, employing negative-design strategies such as charge/size biasing and multistate filtering to prevent off-target assembly.
Structure Prediction and Filtering
AlphaFold2 (AF2) was used to predict and evaluate designed interfaces. Only models with Cα RMSD < 2 Å, mean predicted aligned error (pAE) < 10, and predicted local distance difference test (pLDDT) > 90 for the intended C–D interface and worse metrics for off-target ones—were retained.
3. Experimental Validation
Expression & Assembly
A, B, C, and D subunits were expressed in E. coli, assembled into (ABC)₅ and (ABD)₅ rings, and purified. One design—pD5‑14—assembled correctly into bifaceted structure without forming symmetric off-target aggregates.
Biophysical Characterization
Dynamic Light Scattering (DLS) and Mass Photometry confirmed ~25 nm particles (~1,150 kDa), matching the design (~1,252 kDa).
Negative-stain EM showed monodisperse particles matching the target architecture, and thermal stability testing demonstrated remarkable resilience even up to 95 °C.
High‑Res Structure Confirmation
Cryo‑EM at 4.3 Å resolution matched the design model with with a backbone root-mean-square deviation (RMSD) of ~3.0 Å for the full assembly and ~1.3 Å for the asymmetric C–D interface, confirming high design accuracy.
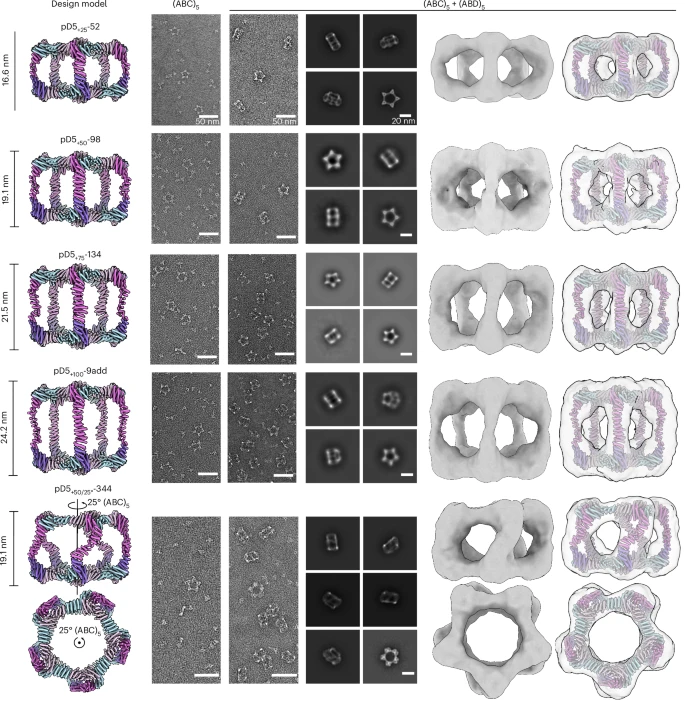
4. Structural Tuning: Variants by Size & Shape
Researchers leveraged RFdiffusion and ProteinMPNN to systematically vary particle size and shape: editing loops in the C subunit and shifting assemblies along the symmetry axis by 25–100 Å, with or without rotation.
The modified particles retained intended assembly, as confirmed by SEC, nsEM, and DLS, demonstrating precise control over overall architecture and aspect ratio.
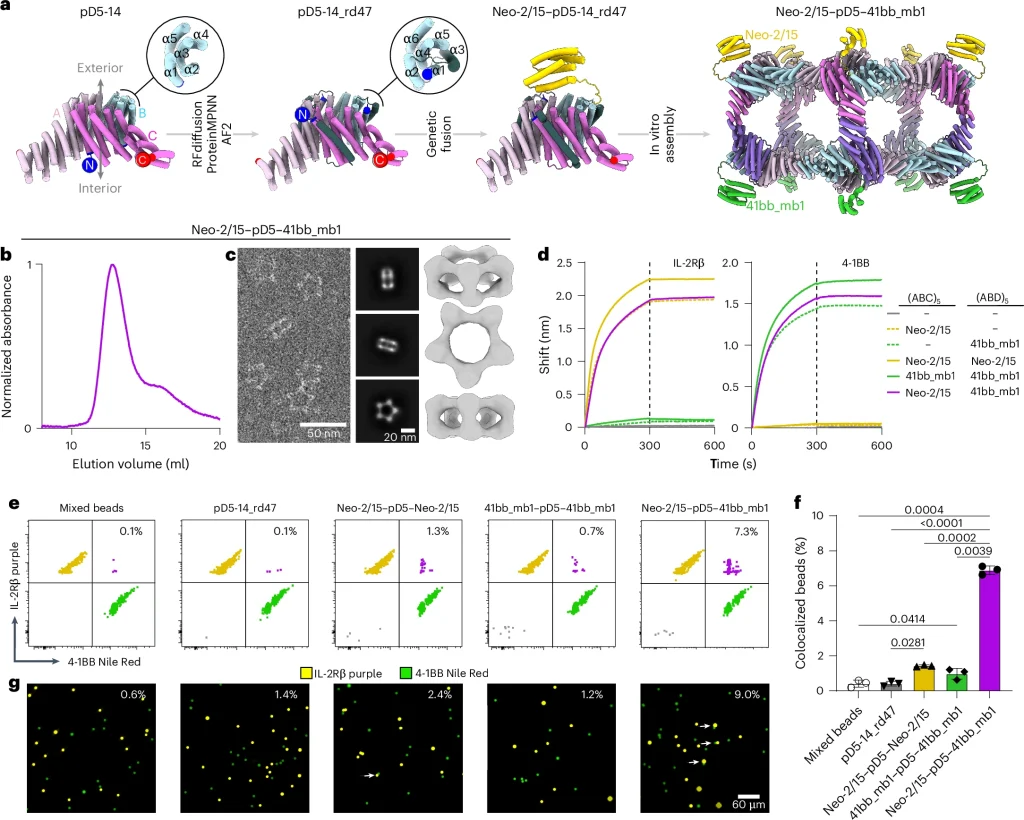
5. Functionalization & Target Colocalization
To enable dual targeting, N‑termini on one face were redesigned (e.g., pD5‑14_rd47) so functional minibinders—targeting IL‑2Rβ and 4‑1BB—could be fused genetically to B subunits.
In colocalization assays, bifunctional nanoparticles significantly increased dual binding events (~6.9%) compared to single-functional or control particles (~1–1.4%), validated via flow cytometry and fluorescence microscopy (~9% vs ~1‑2%).
6. Broader Implications & Future Perspectives
The method introduces a generalizable framework for designing anisotropic, bifaceted proteins with precise structural control, using AI tools such as RFdiffusion, ProteinMPNN, and AF2.
Key advantages:
Independent functionalization of subunits for colocalizing distinct molecular targets.
High thermal resilience, structural precision, and tunability.
Potential applications in dual-antigen display, tissue engineering, immunotherapy, and programmable nanodevices.
This study marks a substantial advance in computational protein nanomaterial design, enabling creation of bifaceted, anisotropic nanoparticles with tunable architectures and targeted dual functionality. The validated pD5-14 assembly demonstrates that AI-driven design can produce highly accurate, experimentally verifiable structures, holding promise for applications in biomedical engineering and molecular therapeutics.
You can find the entire article on the link below: