Amyloid fibrils are central to neurodegenerative diseases like Alzheimer’s and Parkinson’s. However, predicting their structures is notoriously difficult because, unlike most proteins that fold into a single stable form, amyloids exhibit extreme polymorphism: a single sequence can fold into multiple distinct, stable structures depending on conditions. Existing tools like AlphaFold2, designed for monomeric, soluble proteins, struggle with this complexity.
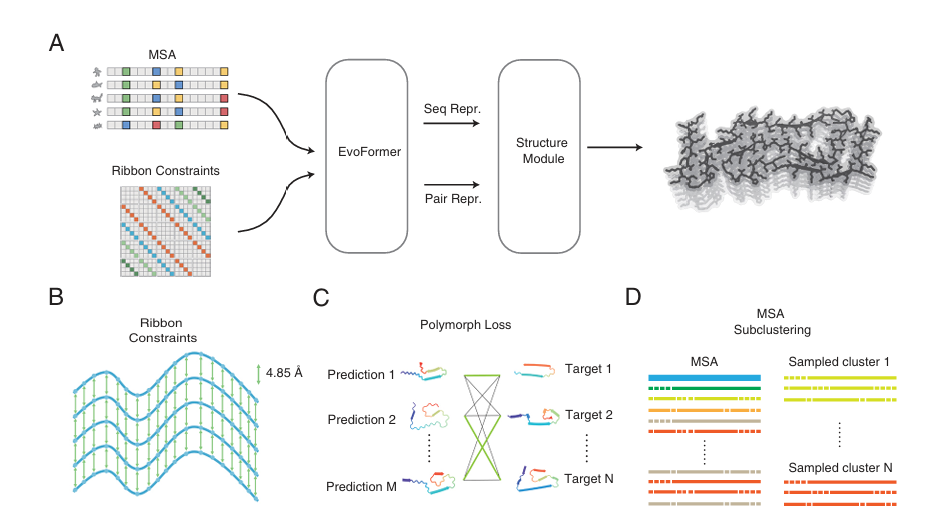
To address this, the study introduces RibbonFold, an AI-driven tool purpose-built to explore the polymorphic landscape of amyloid protofilaments. RibbonFold adapts AlphaFold2’s architecture with key innovations:
Incorporating parallel in-register β-sheet constraints, a hallmark of amyloid structures.
A polymorphism loss function enabling the model to predict multiple plausible amyloid folds for a single sequence.
RibbonFold’s approach is inspired by the “ribbon hypothesis”, treating amyloid fibrils as two-dimensional folding problems constrained by parallel β-sheets, which simplifies the sampling of possible structures.
Key Methodology
Training Data: RibbonFold was trained on 822 curated amyloid ribbon structures clustered into 42 sequence clusters, ensuring a broad sampling of amyloid diversity.
Architecture: Adapted from AlphaFold2-Multimer with specialized encoding of interchain distances (~4.85 Å) for parallel β-sheets.
Polymorphism Loss: For each sequence, RibbonFold generates multiple predictions and compares them with all known polymorphic targets, encouraging exploration of structural diversity.
Sampling Techniques:
Random MSA subclustering to diversify predictions.
MSA column dropout for richer input representation.
Training: Models were trained on 24 NVIDIA A100 GPUs for three days using AlphaFold2’s Model 2 checkpoint.
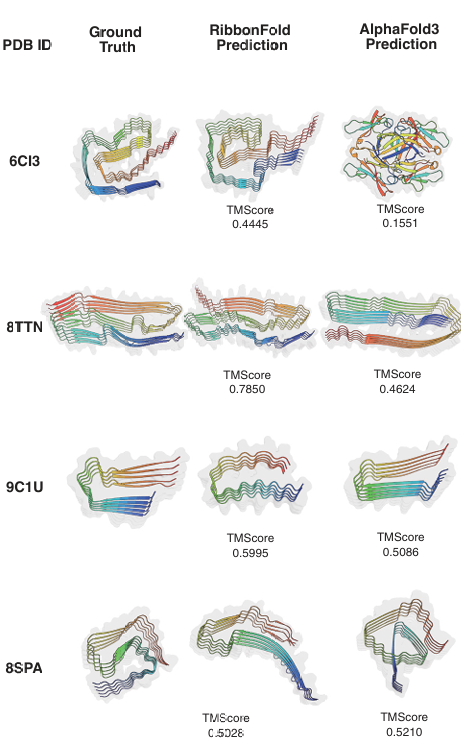
Performance & Validation
Significant Outperformance: RibbonFold outperformed AlphaFold2 and AlphaFold3 on independent amyloid test sets.
Metrics: Mean TM-scores of 0.5 were achieved on test sets, with RibbonFold producing accurate parallel-in-register ribbon-like structures while AlphaFold models tended toward globular predictions.
Ablation Studies: Confirmed critical contributions of parallel constraints and polymorphism loss to the model’s accuracy.
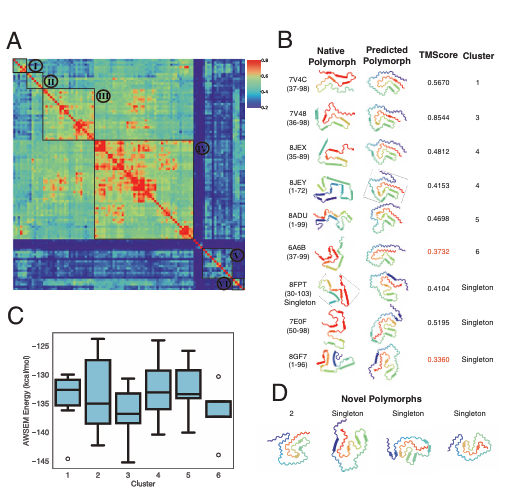
Key Findings
Aβ1-42 Polymorphs: RibbonFold predicted five major polymorphs, with seven of ~10 known experimental polymorphs matching well (TM-score > 0.5). Notably, predicted S-shaped fibrils showed lower solubility and higher stability, aligning with disease-associated forms in Alzheimer’s.
Tau305-379 & α-Synuclein1-99: RibbonFold identified key clusters matching known experimental polymorphs in tau and α-synuclein, central proteins in Alzheimer’s and Parkinson’s, respectively. It also predicted novel polymorphs not yet observed experimentally, providing hypotheses for future studies.
Scrambled Sequences: Predicted structures for scrambled Aβ1-42 sequences were significantly less stable, supporting the idea that disease-associated sequences evolved for aggregation-prone, stable polymorphs.
Implications
RibbonFold reveals that amyloid-forming sequences have a limited but distinct set of energetically favorable polymorphs, and transitions between these states likely require dissolution and re-nucleation due to high kinetic barriers. This insight helps explain amyloid persistence and propagation in neurodegenerative diseases.
Moreover, RibbonFold’s ability to predict novel, plausible polymorphs provides a foundation for therapeutic strategies targeting specific amyloid forms.
Limitations & Future Directions
- Current predictions achieve modest TM-scores (~0.5) due to limited data and the complex polymorphic nature of fibrils.
- Assumes parallel in-register β-sheets; cannot yet predict antiparallel or out-of-register forms.
- Does not account for cofactors or ions critical for in vivo fibril formation.
- Cannot yet model higher-order amyloid fibrils with multiple protofilaments.
You can read the complete article here.